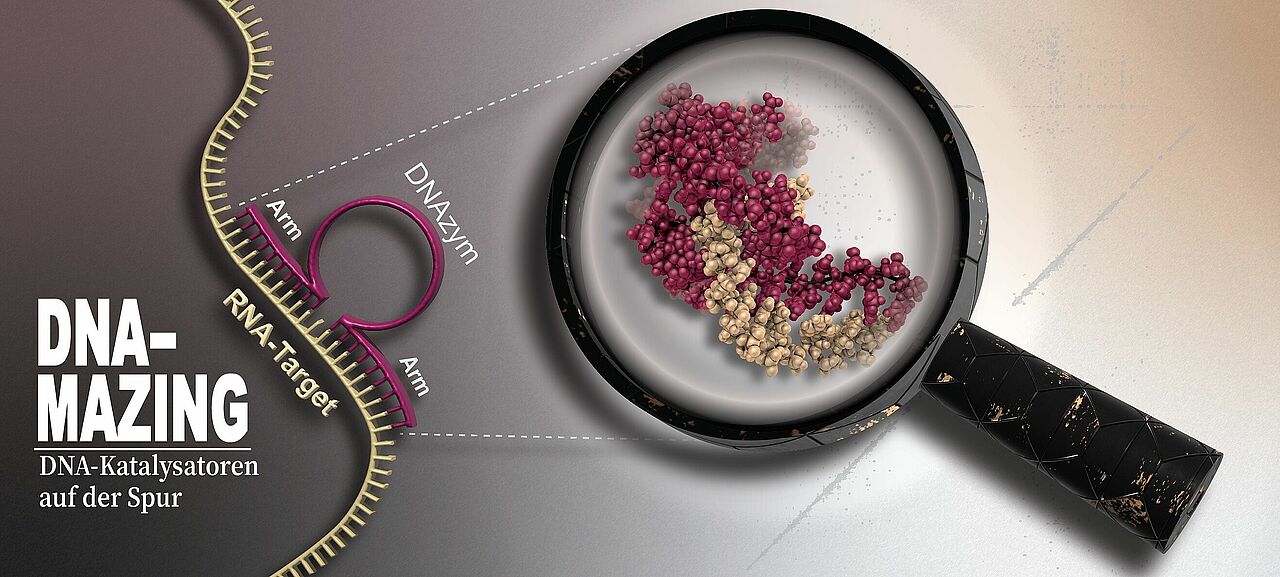
DNAzymes – a word made up of DNA and enzyme – are catalytically active DNA sequences. They comprise a catalytic core comprising around 15 nucleic acids flanked by short binding arms on the right- and left-hand sides, each with around ten nucleic acids. While the sequence of the core is fixed, the binding arms can be modified specifically to match virtually any RNA target sequence.
The aim is to target unwanted RNA molecules of viruses, cancer or damaged nerve cells, using DNAzymes to attack and destroy them. This is achieved via binding sequences that match a sequence of nucleotides on the targeted RNA molecule. The DNAzyme docks precisely to the matching position and the core cleaves the RNA molecule, the fragments of which are then quickly degraded in the cell. The binding arms can be exchanged quickly and easily.
The therapeutic benefits are obvious: Unwanted RNA can be destroyed precisely, while other, useful RNA strands in a cell remain untouched. In some viruses like SARS-CoV2 and Ebola, the genetic material is coded on an RNA molecule. Like healthy cells, cancer cells use so-called messenger RNA (mRNA) to copy the blueprints for proteins from their DNA and transfer them to the molecule factories. The mRNA sequence in cancer cells is often slightly different to that of healthy cells or present in different amounts, meaning that DNAzymes can specifically attack cancer cells while sparing others.
“What sounds outstanding in theory and was already proposed 20 years ago, unfortunately doesn’t work like that in medical practice,” says Dr Manuel Etzkorn, working group leader at the HHU Institute of Physical Biology and last author of the study, which has now been published in Nature. “In a test tube, the DNAzymes are highly effective at destroying the RNA molecules, but this rarely happens in a cell. There must be a competing process that blocks the DNAzymes. However, without a fundamental understanding of how they function, it is very difficult to develop improved DNAzyme variants that can accomplish their work in cells. Our insights have now brought movement into this deadlocked situation.”
In their study, the authors from HHU and a team from Jülich Research Centre (FZJ), the University of Bonn and a Swiss company sought to understand how the system as a whole functions dynamically, what steps occur in the binding and cleaving process and what cofactors support the reaction.
The researchers observed the processes at atomic resolution and in part in real time using high-resolution nuclear magnetic resonance (NMR) spectroscopy. This enabled them to depict the three-dimensional atomic arrangement assumed by the DNAzyme to bind to and cleave the RNA: The core wraps around the RNA strand in a highly effective way, cleaving it into two pieces in several intermediate steps. After cleaving, the DNAzyme releases the fragments and can bind again elsewhere.
Professor Dr Holger Gohlke from the HHU Chair of Pharmaceutical and Medicinal Chemistry and the Institute of Bio- and Geosciences at FZJ, whose team conducted molecular dynamics simulations on the DNAzyme/RNA complex, adds: “In the best sense of integrative modelling, we were able to put forward a plausible RNA cleaving mechanism at atomic level and supply information on RNA base preference at the cleavage site.”
Jan Borggräfe, doctoral researcher in Etzkorn’s working group and lead author of the study, explains why the DNAzymes do not work well in cells: “We established that magnesium, as a key cofactor, plays various essential roles in the mechanism, but that it binds relatively poorly and only briefly to the DNAzyme. There are other components in the cell with a greater affinity for magnesium that “steal” the magnesium from the DNAzyme so to speak.”
The next step is to conduct structural investigations into cell cultures and organoids. The goal for therapeutic applications is to improve the magnesium affinity of the DNAzymes through targeted modifications in order to increase their activity in biological tissue.
Dr Etzkorn states a further area of application: “The focus of our Institute lies on research into neurodegenerative diseases, where we also see good potential for DNAzymes. In the case of Parkinson’s disease, they may under certain circumstances be able to destroy the mRNA sequence that drives the production of alpha-synuclein which, in large quantities, can promote neurotoxic processes.” DNAzymes could also give rise to a new class of antibiotics.
Professor Dr Dieter Willbold, Director of the HHU Institute of Physical Biology and the FZJ Institute for Structural Biochemistry, adds: “The study is yet another example of how basic research in structural biology can provide essential contributions to ground-breaking biomedical advances. The new flagship of the Biomolecular NMR Center, a 1.2 GHz NMR device, has already contributed to this success.” The device at the Biomolecular NMR Center, which is jointly operated by HHU and FZJ, is one of the most powerful systems in the world and provides unique insights into the structure and functioning of the building blocks of life. A short documentary film about the complex installation of the device can be found here.
Original publication
Jan Borggräfe, Julian Victor, Hannah Rosenbach, Aldino Viegas, Christoph G.W. Gertzen, Christine Wuebben, Helena Kovacs, Mohanraj Gopalswamy, Detlev Riesner, Gerhard Steger, Olav Schiemann, Holger Gohlke, Ingrid Span, Manuel Etzkorn, Time-resolved structural analysis of an RNA-cleaving DNA catalyst, Nature (2021).